Abstract
ETP-ALL, an orphan disease, is a subtype of T-ALL with poor prognosis and high risk of relapse after standard of care (SOC) chemotherapy. ETP-ALL is a genomically heterogeneous disease with many cases harboring complex karyotype and genetic mutations typically seen in myeloid neoplasias, such as acute myeloid leukemia (AML). Targeted therapies for ETP-ALL are limited because identified gene mutations are not actionable by currently available drugs. Unfortunately, empirical chemotherapy trials have yet to show significant improvements in clinical outcomes of ETP-ALL patients (pts), which makes treating ETP-ALL challenging. Ideally, selecting treatment for ETP-ALL is rationally designed from the panoply of unique molecular abnormalities observed in ETP-ALL cases. Recently, we developed a predictive simulation modeling method that projects protein network maps from the hundreds of leukemia genomic mutations observed in individual pts (PMID 27855285). Our computational modeling enables multi-gene/multi-drug simulations. We hypothesized that ETP-ALL is comprised of protein network disturbances distinct from other T-ALL cases. We furthermore hypothesized these unique ETP-ALL protein networks would lead to the discovery of new treatment combinations for pts with ETP-ALL.
Aim: Identify novel combinations of drugs for ETP-ALL pts who have failed SOC therapy by using an integrated genomics and computational approach.
Methods: 66 ETP-ALL pts genomic data (whole exome sequencing, copy number variations, cytogenetics) were collected from various sources including NCBI-GEO dataset (GSE28703) (N=12), Moffitt Cancer Center (N=6) (IRB201600284) and published literature (PMID 22237106, 19147408) (N=48). The genomic data for each profile were entered into a computational biology modeling (CBM) software (Cellworks Group), which generated a disease-specific protein network map using PubMed and other online resources. Disease-specific biomarkers were identified within the protein network maps using CBM technology. A digital library of FDA-approved drugs, along with investigational agents, was simulated on every pt model both as single agent and combinations at varying doses. Drug impact was assessed by quantitatively measuring drug effect on a cell growth score (CGS), which is a composite of cell proliferation, viability and apoptosis. Unique combinations that reduced the CGS by interacting with the pt specific disease-biomarkers were selected. Synergy of drug combinations was calculated using the Co-efficient of Drug Interaction (CDI) formula. Each novel therapeutic agent or drug combination was mapped to the pts genomics, and supported by a PubMed referenced scientific mechanism of action based on disease biology.
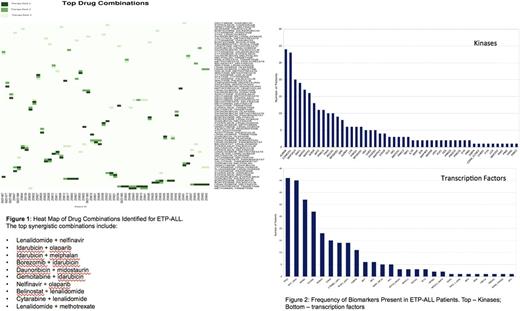
Results: Of the 66 ETP-ALL profiles, 22/66 (33%) harbored actionable mutations, including NRAS, TET2, FLT3 and various deletions such as del(5q) or deletions in tumor suppressor genes including CDKN2A / 2B, TSC1 and others. 44/66 (67%) had many deleterious mutations that were not directly actionable by currently available drugs. The integrated genomics and CBM workflow identified targeted combination therapies for 59/66 (89.4%) of cases. Eighty-seven unique targeted combinations were identified across the 66 ETP-ALL pts in this study (Fig 1). Eleven of the identified combinations were synergistic, as calculated by CDI, and 5 of the combinations included idarubicin as one of the agents, while the most synergistic combination was lenalidomide + nelfinavir (CDI = 3.34). Analysis of CBM identified pt-specific biomarkers for 66/66 (100%) pts. Dysregulated protein networks unique to ETP-ALL pt cases included pathways involving TP53, MYC, NFKB1, FOXM1, RUNX3, STAT3, CTNNB1, E2F1 and CEBPA. Some of the key kinase biomarkers identified in the ETP-ALL profiles were AURKB, CSNK2A1, MAP3K14, RAF1, ERK and mTOR (Fig 2).
Conclusions: Computational biology analysis of complex ETP-ALL genomics found a unique protein network that may be amenable to several new treatment combinations. This CBM approach identified personalized drug regimens in all cases, including those who had no actionable genetic mutations present. This predictive technology improves the utility of cancer genetic profiling and may enhance clinical decision-making to identify treatments for ETP-ALL pts who would otherwise have no options based on limited SOC alternatives.
Kumar: Cellworks: Employment. K: Cellworks: Employment. Bhargav: Cellworks: Employment. Choudhary: Cellworks: Employment. L: Cellworks: Employment. Birajdar: Cellworks: Employment. Roy: Cellworks: Employment. Abbasi: Cellworks Group Inc.: Employment. Vali: Cellworks Group Inc.: Employment. Cogle: Celgene: Other: Membership on Steering Committee for Connect MDS/AML Registry.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal